2016-2019
Engineering Metabolism
A partial C4 pathway was successfully introduced into rice using a single construct containing 5 genes encoding C4 enzymes (four expressed in the mesophyll and one in the bundle sheath). After development of suitably sensitive methods to measure levels of the four carbon compound malate, carboxylation by PEP carboxylase was detected in the form of higher levels of malate in transgenic lines as compared to wild-type. However, subsequent decarboxylation by malic enzyme could not be detected, with 3-PGA dynamics being the same in both wild-type and transgenic lines (see paper here). This observation initiated the search for more effective regulatory sequences for the activation of decarboxylases in rice bundle sheath cells, and also prompted a series of studies in both Setaria viridis and rice to better understand what might be limiting pathway function, including a focus on metabolite transporter proteins.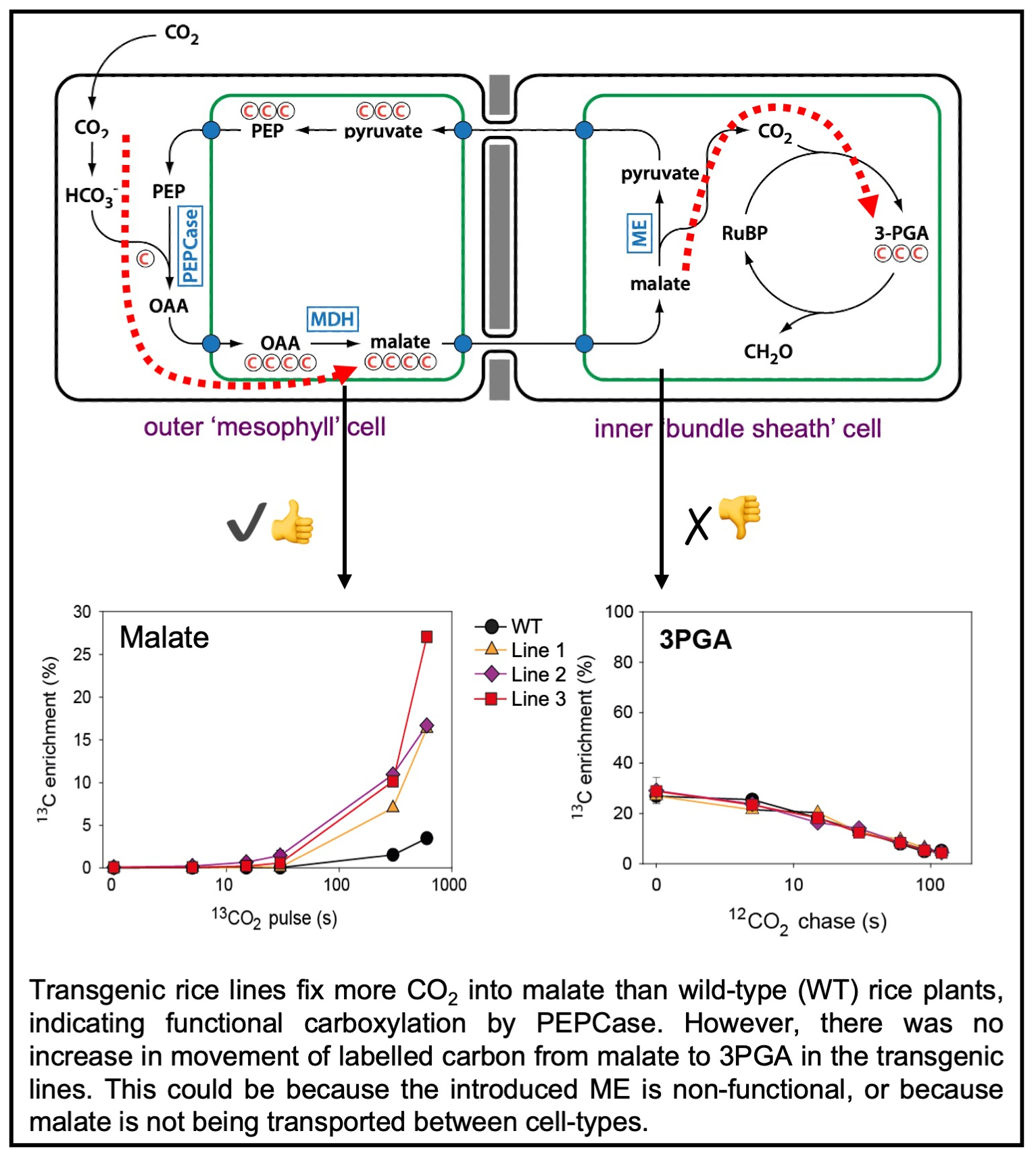 Engineering Anatomy
Engineering Anatomy
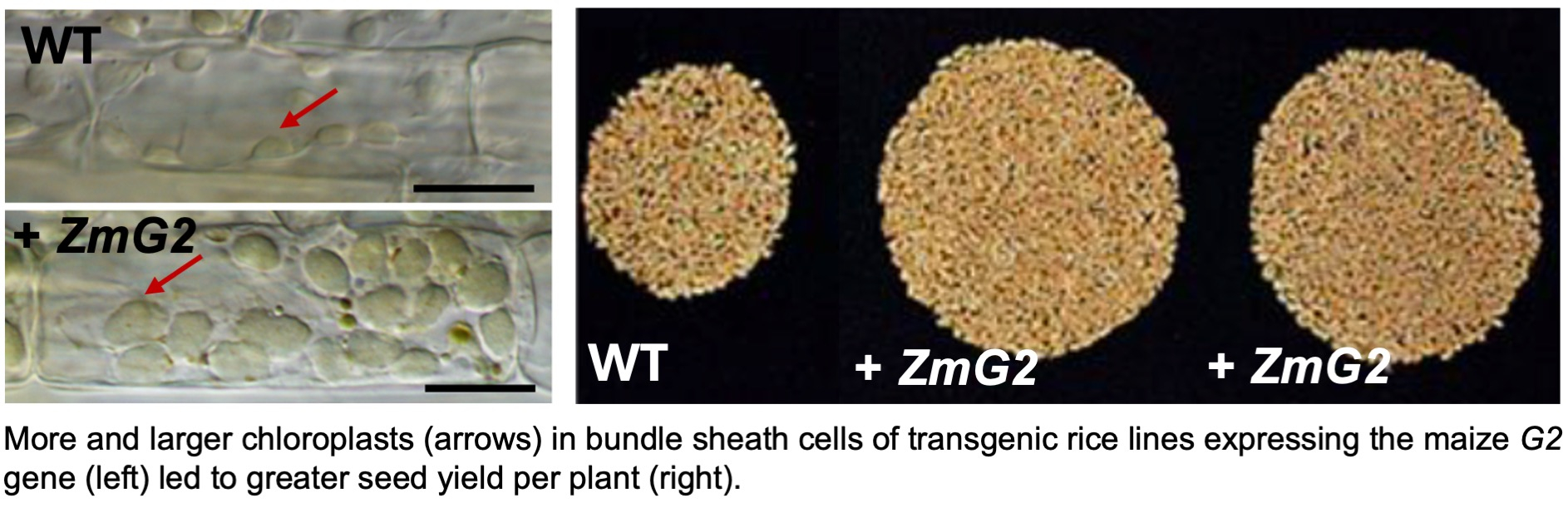
The search for regulators of vein spacing from within the list of candidate regulators of C4 leaf anatomy was difficult, with little information available about the roles of any of the candidate genes in other species. A brute force strategy was therefore adopted, analysing the effects of constutively activating the maize genes in rice (IR64 and Kitaake). Although far less than an ideal approach, 60 putative regulators were eliminated. Published in 2017 (see paper here), this study reduced the number of candidate genes to around ten. One of the remaining genes encoded the GRAS transcription factor SHORTROOT (SHR), which was known to play a role in radial patterning of cell-types in the Arabidopsis root. Much has been published on the role of SHR in grass leaf development but the data are conflicting and confusing. In our hands, constitutive overexpression in rice was not tolerated, in that seedlings would not survive after regeneration from transformed callus. However, by ectopically expressing the maize SHR gene preferentially in rice bundle sheath cells, extra stomatal files developed on the leaf surface (see paper here). Notably, this is a trait that will ultimately be needed in C4 rice and it was therefore reassuring to see that the increased stomatal density in itself had no negative impact on photosythesis. Whereas regulators of vein spacing remained elusive, the maize transcription factor GOLDEN2 (G2) (and its orthologs) were known to regulate chloroplast development in all plant species examined. With a view to activating chloroplast development in rice bundle sheath cells, the maize G2 gene was therefore constitutively expressed in rice. Surprisingly, transgenic lines not only exhibited activated chloroplast development in bundle sheath cells but also an increase in mitochondrial and plasmodesmatal numbers in that cell-type. Emergent traits after the manipulation of one aspect of anatomy was becoming a theme. This major advance was published in 2017 (see paper here). Although chloroplast volume in the bundle sheath cells of transgenic lines overexpressing the maize G2 gene did not match that seen in C4 plants, field experiments carried out with collaborators at the Chinese Academy of Agricultural Sciences revealed a > 20% yield increase, albeit in the Kitaake background rather than an elite variety used for food production (published in 2020 see paper here). With these exciting results, the search for additional regulators that might further increase chloroplast volume and yield was started.
Whereas regulators of vein spacing remained elusive, the maize transcription factor GOLDEN2 (G2) (and its orthologs) were known to regulate chloroplast development in all plant species examined. With a view to activating chloroplast development in rice bundle sheath cells, the maize G2 gene was therefore constitutively expressed in rice. Surprisingly, transgenic lines not only exhibited activated chloroplast development in bundle sheath cells but also an increase in mitochondrial and plasmodesmatal numbers in that cell-type. Emergent traits after the manipulation of one aspect of anatomy was becoming a theme. This major advance was published in 2017 (see paper here). Although chloroplast volume in the bundle sheath cells of transgenic lines overexpressing the maize G2 gene did not match that seen in C4 plants, field experiments carried out with collaborators at the Chinese Academy of Agricultural Sciences revealed a > 20% yield increase, albeit in the Kitaake background rather than an elite variety used for food production (published in 2020 see paper here). With these exciting results, the search for additional regulators that might further increase chloroplast volume and yield was started.