2020-2024
Engineering Metabolism
Systems to increase transgene expression in bundle sheath cells were identified via two routes. In the first, characterization of transcriptomes in bundle sheath cells of rice revealed the importance of sulphur metabolism in that cell-type (see paper here), subsequently leading to the identification of an extremely efficient promoter (OsSiR) for driving bundle sheath cell-preferential gene expression (see paper here). In the second, a synthetic transcription factor/promoter system (dTALE/STAP) was developed for use in rice that enabled a single promoter to activate and enhance the expression of multiple transgenes (see paper here). Both of these components are incorporated into current construct designs, that include genes encoding both C4 enzymes and transporter proteins. One of these transporter proteins is the chloroplastic 2-oxoglutarate/malate transporter which when introduced as a single transgene in rice caused growth to be stunted (see paper here). Biochemical characterization of these lines revealed perturbed nitrogen homeostasis, highlighting the need to monitor crosstalk between carbon and nitrogen metabolism in transgenic lines designed to alter photosynthesis. The presence/absence of suberin in the walls of rice bundle sheath cells will also need to be considered in any final C4 rice line, given that characterization of mutants in Setaria viridis revealed the importance of suberized walls for optimized C4 function (see paper here). Rice lines expressing 15 transgenes are currently being evaluated for photosynthetic function.
In parallel to transgenic experiments, gene editing was performed to relocate carbonic anhydrase (CA) from the chloroplast to the cytoplasm of rice mesophyll cells (see paper here). Having shown that reduced levels of CA in Setaria viridis limit C4 photosynthesis (see paper here), it was predicted that extra CA would need to be introduced into rice mesophyll cells once CA was eliminated from the chloroplast. Surprisingly, however, the edited CA lines exhibited cytoplasmic CA activity levels similar to that seen in C4 plants. The first step in the C4 pathway is thus operational in these gene edited lines, providing a chassis into which subsequent steps of the pathway can now be introduced.
Engineering Anatomy
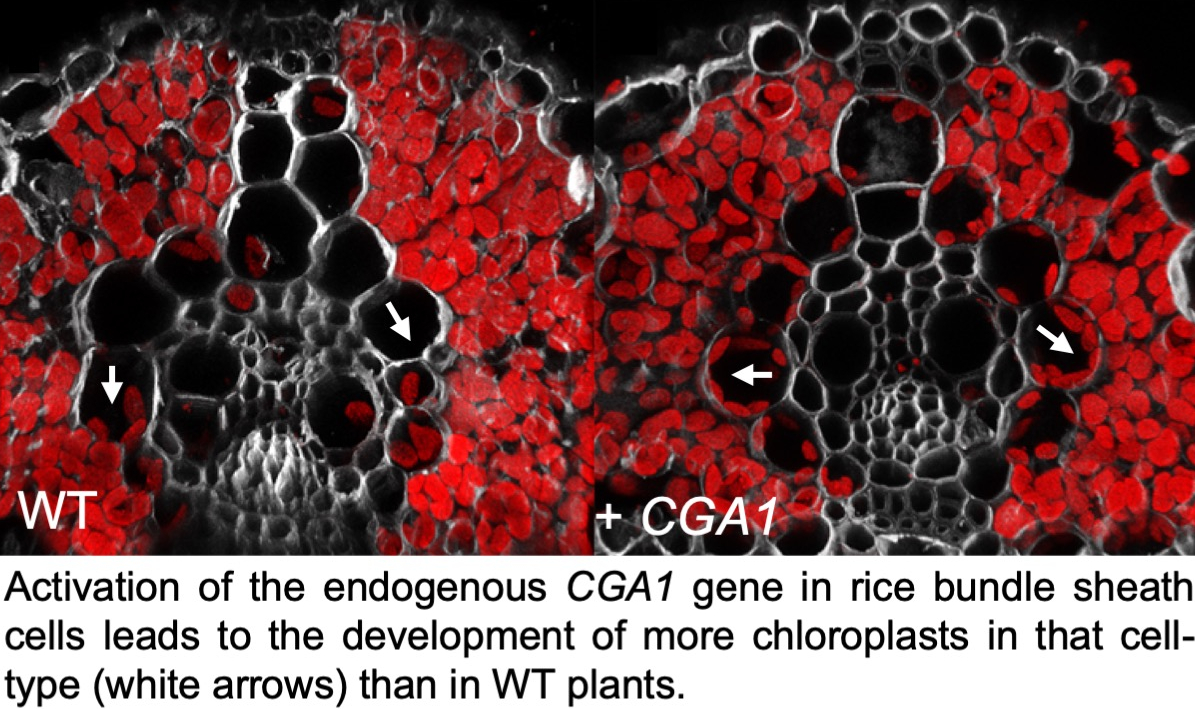
Further analysis of SHORTROOT pathway function, specifically in relation to function of the interacting SCARECROW (SCR) protein, confirmed a role for the pathway in patterning mesophyll cells in the maize leaf (see paper here), and again showed relationships between patterning of inner leaf tissues and stomatal development (see paper here). Unfortunately, however, manipulation of function in rice did not alter inner leaf anatomy but instead abolished stomatal development (which was not very helpful!) (see paper here). Meanwhile, the pursuit for regulators of vein formation continued, resulting in the identification of the zinc finger protein ‘TOO MANY LATERALS (TML)’. The discovery that the TML1 gene is expressed in developing minor but not major veins of maize leaves (see paper here), led to the hypothesis that loss of gene function might influence vein patterning. In an exciting advance, gene edited tml1 lines in both maize and rice were shown to develop more major veins in the leaf than wild-type siblings (see paper here). In rice leaves this results in an overall increase in both vein and bundle sheath cell volume. Although vein spacing is not altered in these lines, the increased bundle sheath cell volume provides space to further enhance bundle sheath chloroplast volume above levels possible in wild-type plants. The tml1 gene edited line thus provides an anatomical chassis into which bundle sheath chloroplast development can be manipulated prior to the introduction of C4 metabolism.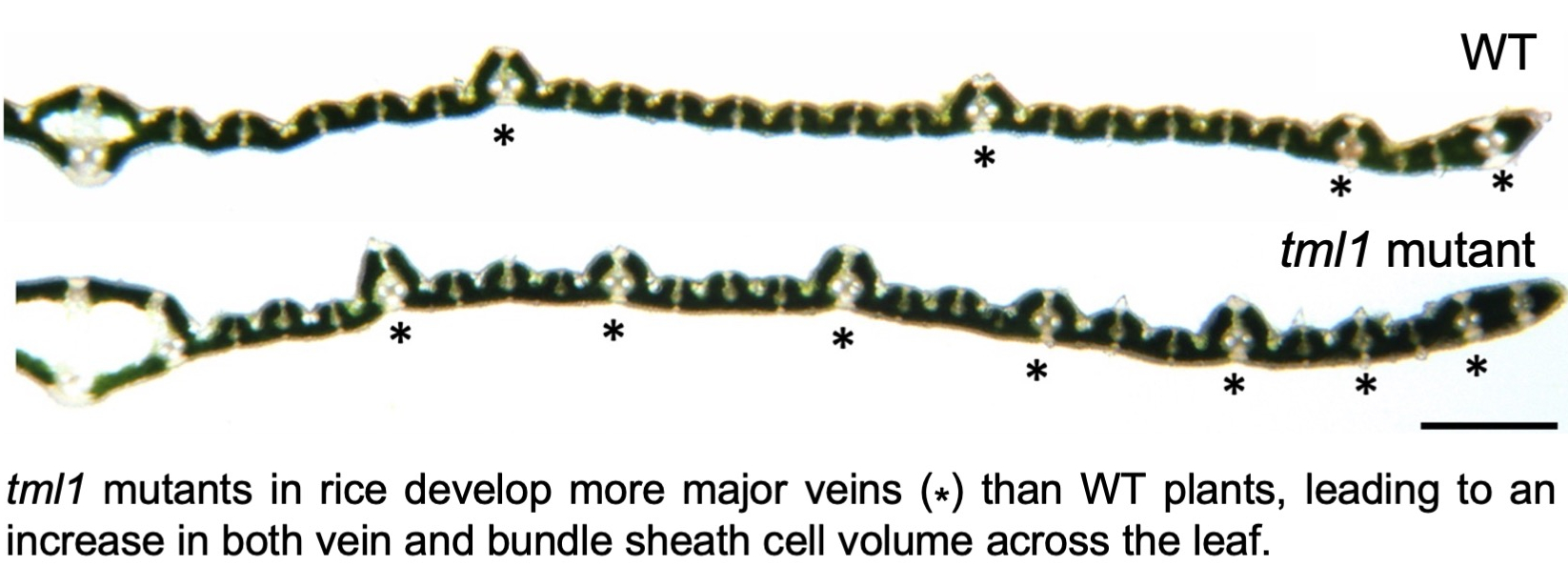 The search for additional activators of chloroplast development continued, requiring the establishment of rapid (see paper here) and/or deep-learning based (see paper here) methods to quantify chloroplast volume in the process. Although not as great as that seen in lines overexpressing the maize G2 gene, an increase in bundle sheath chloroplast content was observed in lines in which the rice CGA1 gene was activated in bundle sheath cells (see paper here). These observations prompted experiments to simultaneously manipulate both genes, the tantalizing results of which are currently being evaluated.
The search for additional activators of chloroplast development continued, requiring the establishment of rapid (see paper here) and/or deep-learning based (see paper here) methods to quantify chloroplast volume in the process. Although not as great as that seen in lines overexpressing the maize G2 gene, an increase in bundle sheath chloroplast content was observed in lines in which the rice CGA1 gene was activated in bundle sheath cells (see paper here). These observations prompted experiments to simultaneously manipulate both genes, the tantalizing results of which are currently being evaluated.