2009-2015
Engineering Metabolism
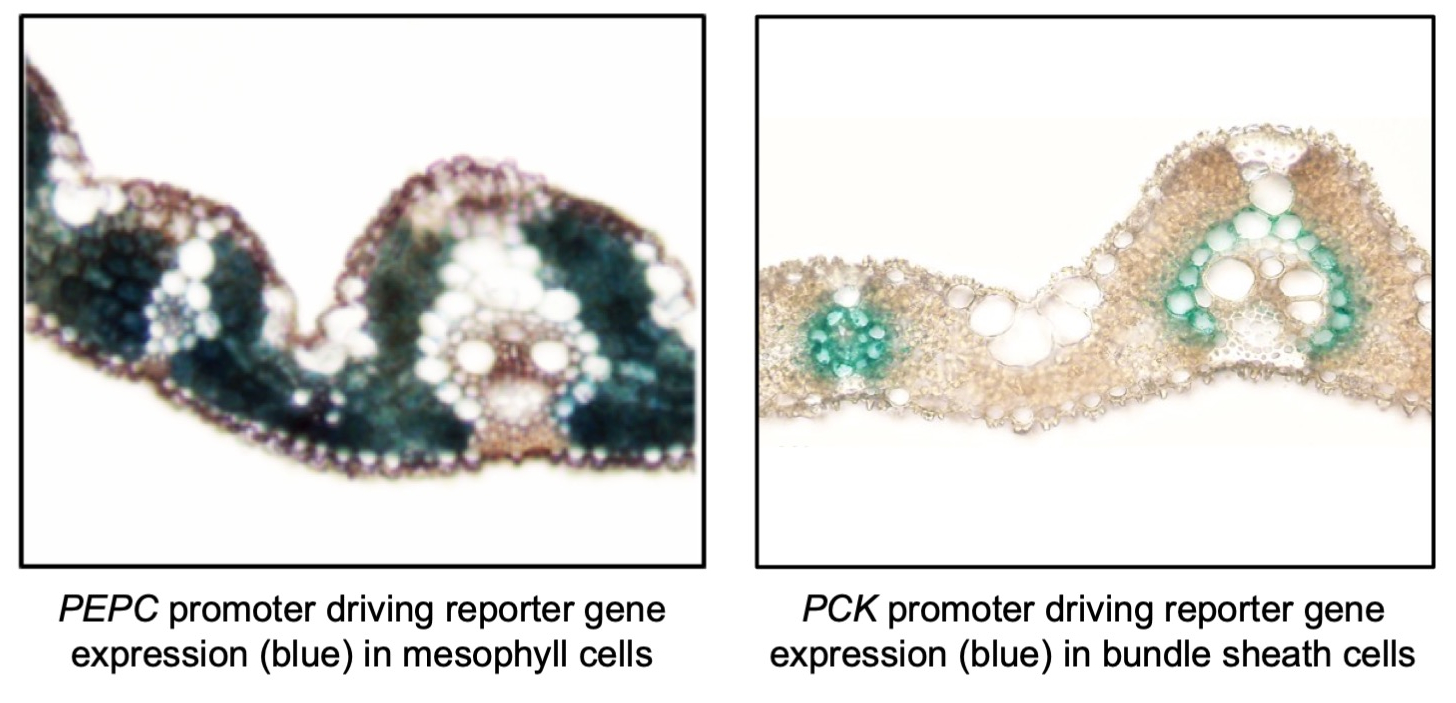
To initiate the work on engineering metabolism, protocols were optimized for the introduction (‘transformation’) of genes into the indica rice variety IR64 and DNA sequences were identified that switched on genes (‘promoters’) in either mesophyll or bundle sheath cells of rice. Notably, most of the promoter sequences that activated cell-preferential gene expression in C4 species, did not replicate that pattern in rice and so the search was long and hard. However, by 2015 a number of promoters that functioned in rice mesophyll cells had been identified and two had been identified that functioned (albeit weakly) in bundle sheath cells.
These advances allowed individual genes encoding C4 enzymes to be introduced into IR64, with gene activation driven by the appropriate cell-specific promoter – four genes activated in the mesophyll cells and one in the bundle sheath cells. Genetic crosses began to combine the 5 C4 genes into a single line, a process that took 7 years to complete. Fixation of carbon dioxide into C4 acids was detected in the ‘stacked’ lines but with each of the 5 transgenes inserted into a different region of the genome, the lines were inherently complex and difficult to work with (published in 2020 see paper here). In 2014, synthetic modular cloning was therefore adopted across the consortium to facilitate the rapid generation of constructs containing multiple genes that could be inserted into a single region of the genome. Notably this required the redesign of many cloning modules. For example, commonly used fluorescent reporter proteins could not be detected above levels of autofluorescence in rice and thus modules had to be re-designed for use specifically in rice see paper here. To further speed progress, a transformation pipeline was established that exploited the rapid cycling japonica rice variety ‘Kitaake’, allowing seed to seed generation in 5 months and faster evaluation of multigene constructs. This development expanded the capacity for transformation experiments to four of the consortium labs.
Engineering Anatomy
To determine the extent to which vein spacing patterns can and/or do vary in grass leaves, large scale forward mutant screens in sorghum (C4) and rice (C3) (see paper here) were carried out, along with analyses of wild rice relatives see paper here. Notably, little variation was observed suggesting that neither forward genetics or natural variation was likely to deliver a C4-like leaf anatomy. Two sorghum mutants were identified with altered vein patterning but both converted leaf blade tissue into leaf sheath rather than altering vein patterning per se (see paper here). Research therefore pivoted both to exploit new genome-wide transcriptomics approaches to identify potential regulators of leaf anatomy, and to establish a C4 model species that could be more easily transformed than maize or sorghum. Transcriptomic analysis of genes expressed during the development of maize foliar (Kranz) and husk (no Kranz) leaves identified the first putative regulators of C4 leaf anatomy (see paper here), and Setaria viridis was adopted as a C4 model for mutant screens and transformation. Before investing significant time and effort into functional analyses of candidate regulators, however, it was necessary to accurately identify orthologous genes in maize, sorghum, rice and setaria. This requirement led to the development of Orthofinder software, and to a paper that has been cited over 2000 times (see paper here).